CGRM: Cardiac gene regulary modelling
What is CGRM?
Heart disease is usually due to a deficiency or dysfunction of cardiomyocyte (CM) or other heart cells in human hearts. The development of heart cells involves complex alterations in gene expression over time, which ultimately contribute to heart formation and function. These developmental processes are governed by a set of transcription factors (TFs), microRNAs (miRNAs), and long noncoding RNAs (lncRNAs), which modulate diverse transcriptional and post-transcriptional activities leading to cardiac commitment, differentiation and maturation. To dissect such complex interactions of multiple regulators, we developed an integrated platform for cardiac gene regulatory modeling, CGRM, that can coordinate heterogeneous cardiac data for comprehensively identifying heart regulatory target programs of TFs, miRNAs and lncRNAs at different developmental stages of heart cells and tissues. The web-based method demonstrates its capability to reveal multi-level gene regulatory networks (GRNs) underlying dynamic cardiac developmental progression.
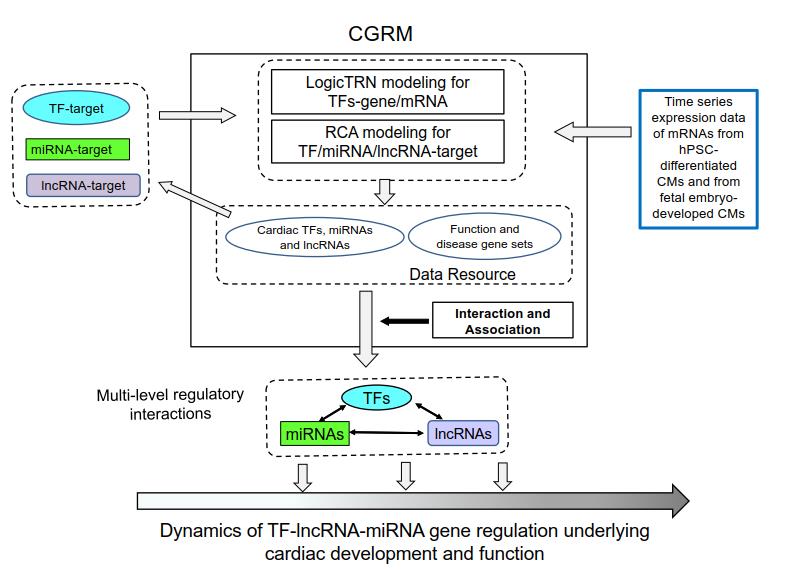
The Figure shown here displays the schematic procedure and an overview layout of CGRM. Basically, the platform comprises two main modeling systems, whose aim is to decode complex interactions driven by the multiple level regulatory factors and to construct the GRNs. The LogicTRN model identifies the logics of TFs and the underlying transcriptional networks by combining time series mRNA expression profiles and TF-DNA binding data (Reference 1). The RCA model identifies the most likely regulatory interaction modules between the factors and targets through matrix decomposition and factorization (Reference 2). The outputs obtained from the two models provide a base to define the regulatory interrelationships between TF-miRNA-lncRNA and targets. The platform also has built-in software and statistical tools to evaluate the enrichment of overlap among the target gene sets of different regulatory factors between the target gene sets and function gene sets, and seek their associations with heart development, biological functions, and heart disease. Alternatively, CGRM enables researchers to conduct new analyses by submitting their own data. The detailed procedure for using CGRM, such as selecting input data and the modeling parameters, can be found on the tutorial page .
References
- Yan B, Guan D, Wang C, Wang J, He B, Qin J, Boheler KR, Lu A, Zhang G, Zhu H: An integrative method to decode regulatory logics in gene transcription. Nature Communications 2017, 8:1044.
- Yan B, Li H, Yang X, Shao J, Jang M, Guan D, Zou S, Van Waes C, Chen Z, Zhan M: Unraveling regulatory programs for NF-kappaB, p53 and microRNAs in head and neck squamous cell carcinoma. PLoS One 2013, 8:e73656.